In genome-wide association study (GWAS), we aim at detecting single nucleotide polymorphisms (SNPs) that are associated with diseases. Conventional SNP interaction detection methods like PLINK [1] and BOOST [2] are based on SNP-SNP interactions. Although single nucleotides are the building blocks of human genome, SNPs are not necessarily the smallest functional unit for complex phenotypes. Region-based strategies have been successful in studies aiming at marginal effects (the effect of a single SNP regardless of other SNPs). In this paper, we have developed RRIntCC [3], a novel region-region interaction detection method for case-control studies, by utilizing the correlations between individual SNP-SNP interactions based on linkage disequilibrium (LD) contrast test.
The test statistic of LD contrast test for two SNP A and B is,
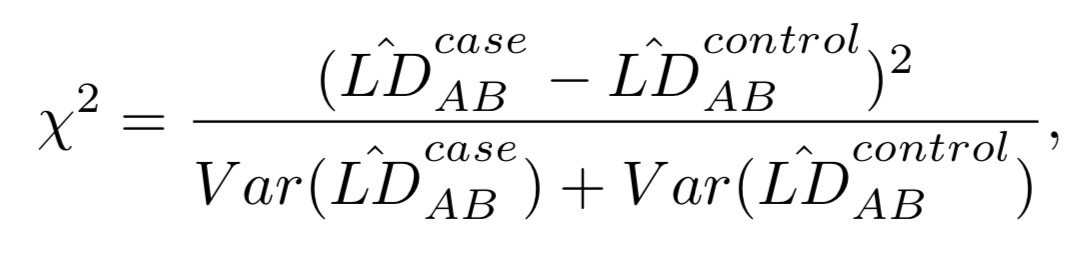
which follows a 1-df chi-square distribution under the null hypothesis that there is no LD difference between cases and controls.
In RRIntCC, we adopted a minimum p-value based method to aggregate SNP-SNP interactions within two regions. The p-value for a region-region interaction reads,
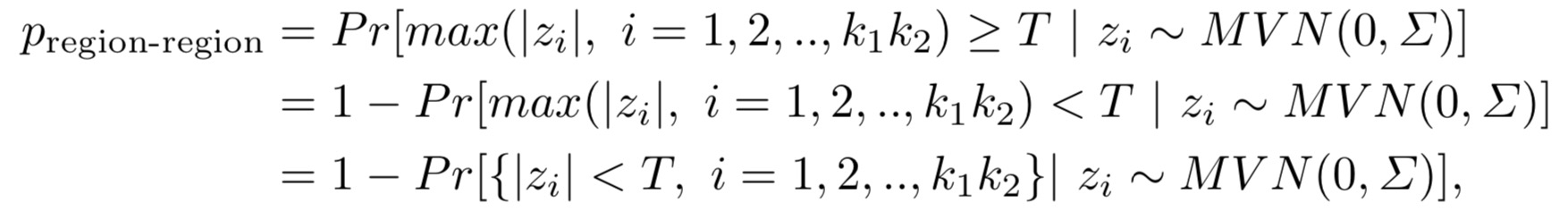
where
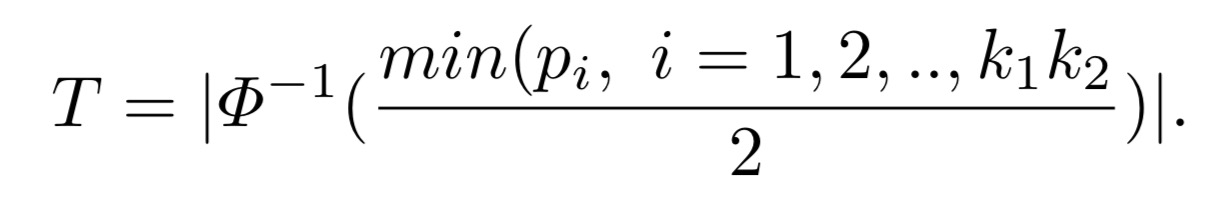
The key contribution of RRIntCC is that we have derived the covariance between the LD contrast test statistics of SNP-SNP interactions so that we can avoid inflated p-values and false positives. Please refer to our RRIntCC paper for more detail.
The source code is available at:
RRIntCC: RRIntCC.zip
Install R, Rcpp and RInside dependencies:
1. Need to install R base env: sudo apt install r-base-core
2. Need to install mvtnorm, Rcpp and RInside (The packages can be downloaded online): sudo R CMD INSTALL xxx.tar.gz
Please read rrintcc_read.me in the zip file for detail.
For any enquiry, please contact ZHANG Sen at szhangat@connect.ust.hk
1. Purcell, S., et al.: PLINK: A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics 81(3) (2007) 559-575
2. Wan, X., et al.: BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. The American Journal of Human Genetics 87(3) (2010) 325-340
3. Zhang, S., et al.: Region-based interaction detection in genome-wide case-control studies. The 14th International Symposium on Bioinformatics Research and Applications, Beijing, June 8-11 (2018)